Andy Jones, Will Townes, Didong Li, and Barbara Engelhardt introduce Gaussian process spatial alignment (GPSA), a probabilistic model that aligns a set of spatially-resolved genomic and histology slices onto a known or unknown common coordinate system.
Spatially-resolved genomic technologies enable the study of the organization of cells and tissues, and promise to explain local interactions between cells. But, it is difficult to precisely align spatial observations across slices, samples, scales, individuals, and technologies.
Given a set of spatial genomics tissue slices with distorted or unaligned local coordinate systems, GPSA estimates a common coordinate system (CCS) as a collection of latent variables from these disparate spatially-resolved measurements and aligns new slices to this estimated CCS
Similar approaches exist. PASTE’s optimal transport framework finds mappings between adjacent slices, but is limited to linear warpings. Eggplant maps slices onto a template coordinate system, but requires landmark annotations.
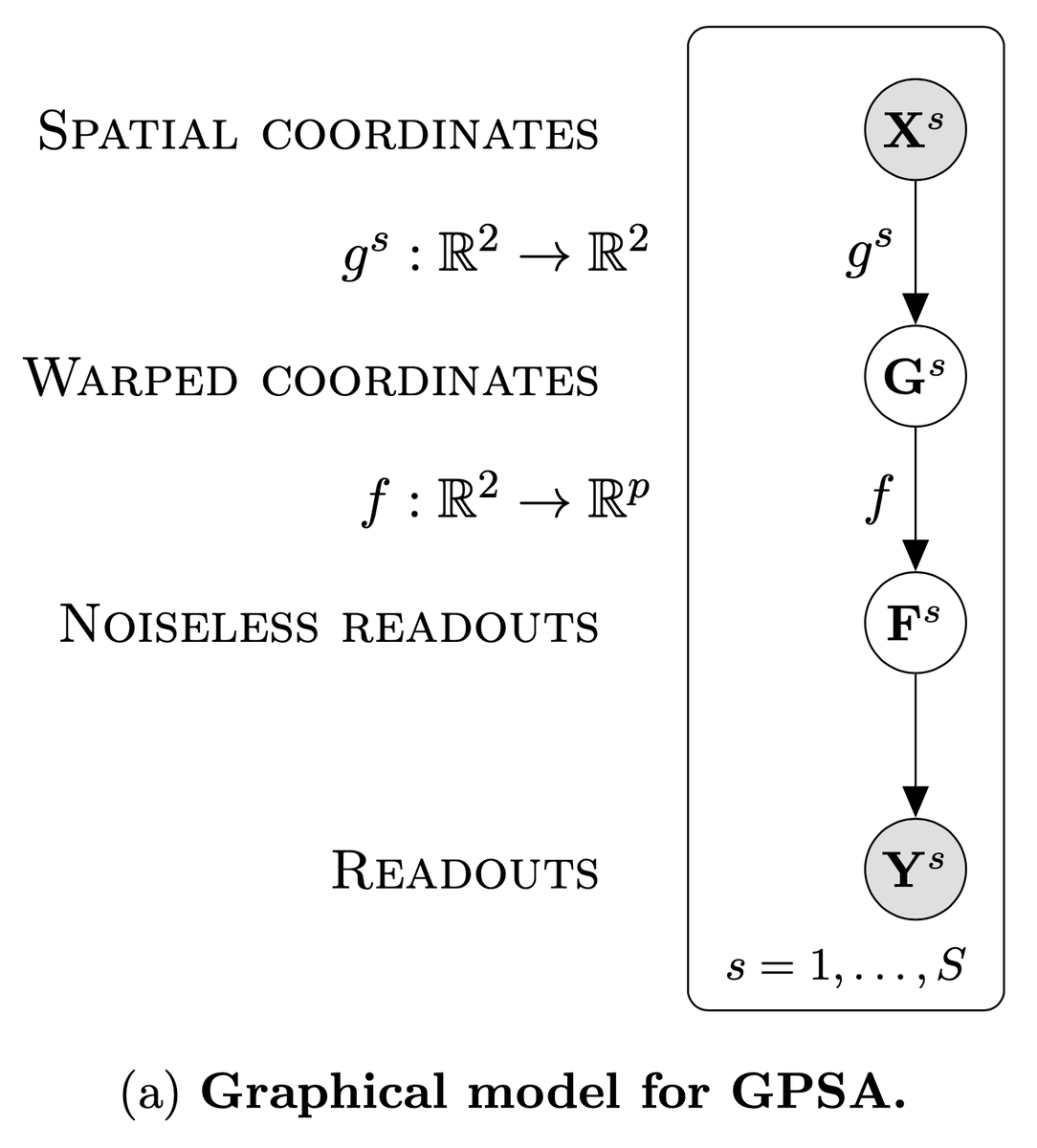
Our GPSA consists of a two-layer deep GP: the first layer maps observed spatial locations into a common coordinate system (CCS), and the second layer maps from the CCS to observed phenotypic readouts. We use variational inference with inducing points for scalable inference.
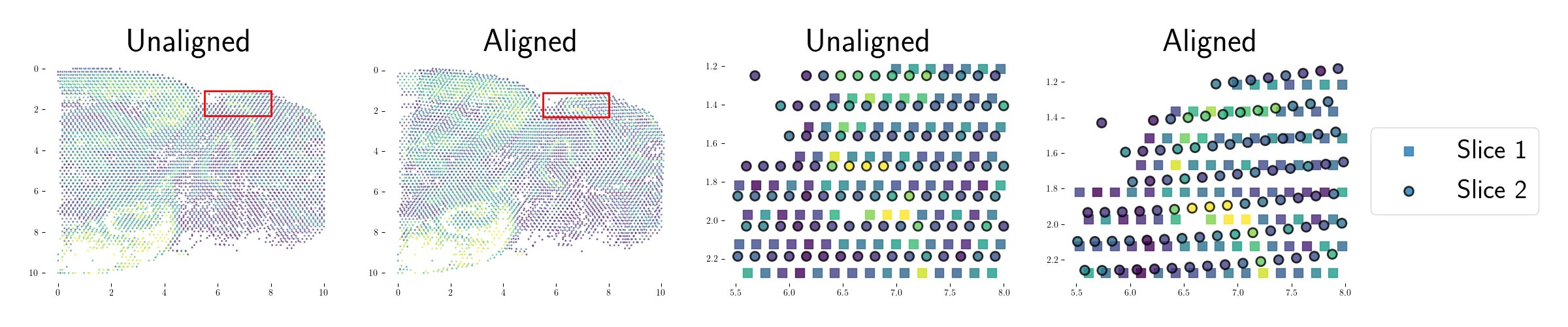
We apply GPSA to a number of spatial genomics technologies (Visium, Slide-seqV2, ST), and find that it can flexibly and robustly model and correct for spatial distortions and biological variability across varying resolutions, fields of view, and modalities.
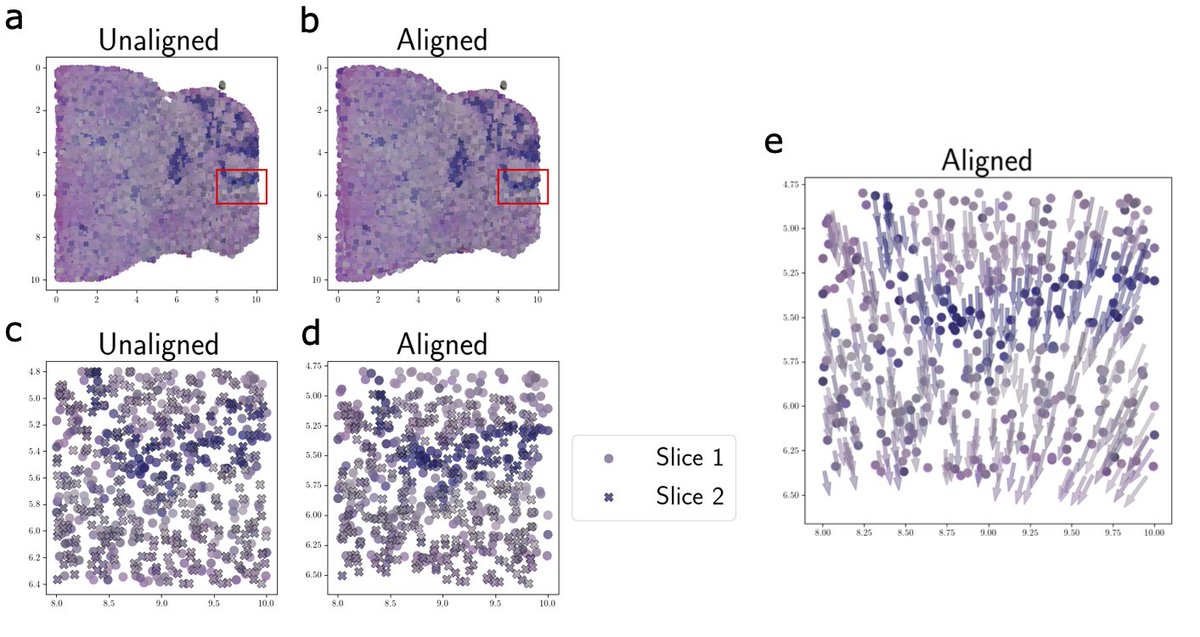
GPSA jointly aligns spatial genomics and histology images, allowing for cross-modality analysis and paving the way for building multi-modal tissue atlases from scratch. Each 3D coordinate in the CCS can be queried for the distribution of the molecular phenotypes at that spot.
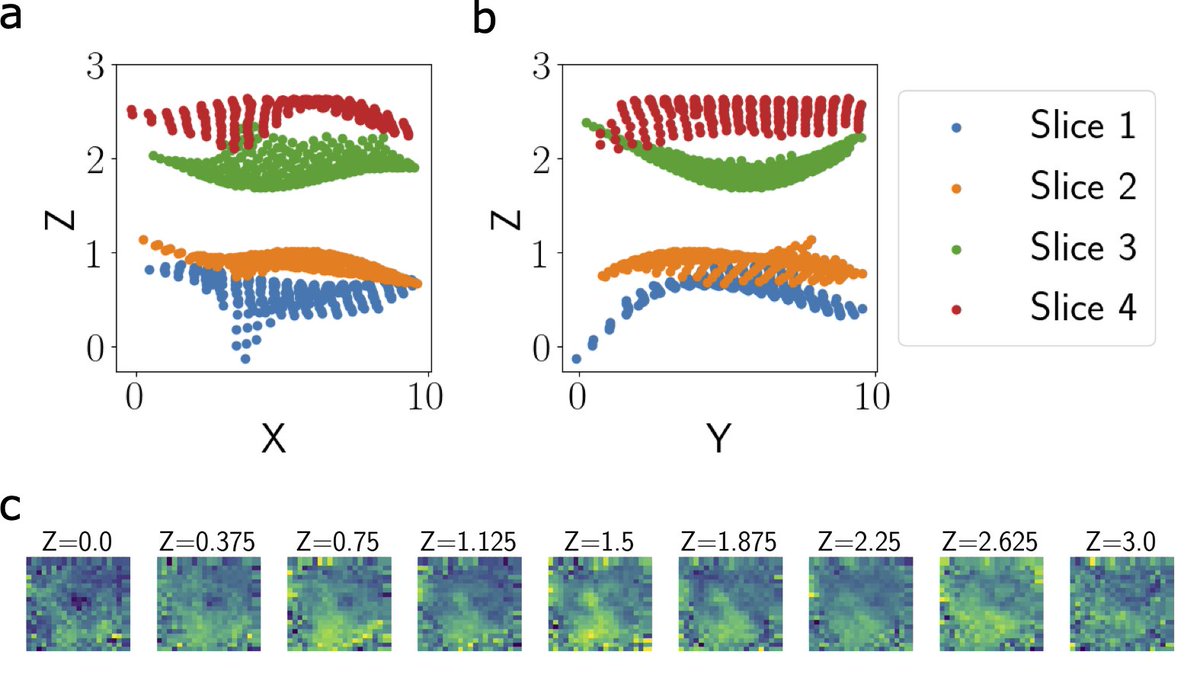
Given a set of parallel 2D tissue slices from the same tissue, GPSA can align these slices into a 3D common coordinate system. This captures phenotypic variation along the z-axis, as we demonstrate with ST data from four adjacent slices of a breast tumor.
GPSA can perform two types of alignments: template-based alignments map slices to an existing coordinate system, and de novo alignments create a new common coordinate system from a set of tissue slices.
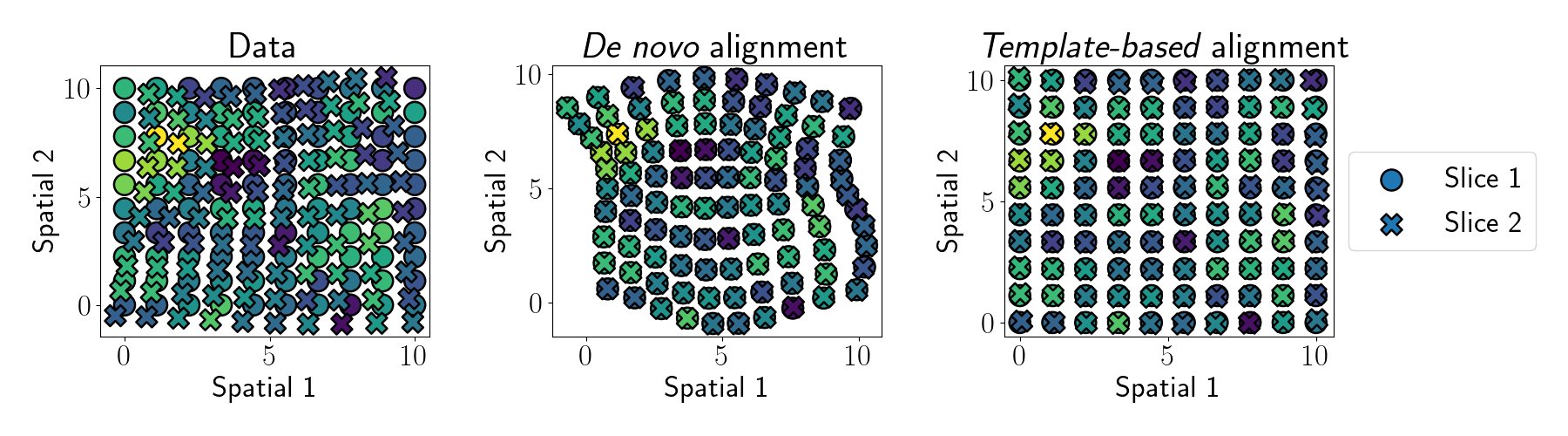
We envision GPSA being useful for a variety of multi-slice downstream tasks. E.g., identifying spatially variable genes, exploring low-dimensional structure across multiple samples, and creating atlases of spatial genomics data for entire tissues or organisms.
Feedback welcome! Try our GPSA Python package to aligning slices of your favorite tissue: [Code]