Poisson reduced rank regression (PRRR)
Tiana Fitzgerald, Andy Jones, Barbara Engelhardt
Our paper developing count-based model for association mapping published in #BMCBioinformatics! Great work by amazing Princeton undergrad Tiana Fitzgerald and PhD student Andy Jones.
A Poisson reduced-rank regression model for association mapping in sequencing data
We develop a Poisson reduced-rank regression model to identify low-dimensional associations in high-dimensional data.
We adapted association mapping methods developed for bulk sequencing to count-based single-cell technologies. The goal here is to identify associations between cell-specific covariates and each cell’s expression levels.
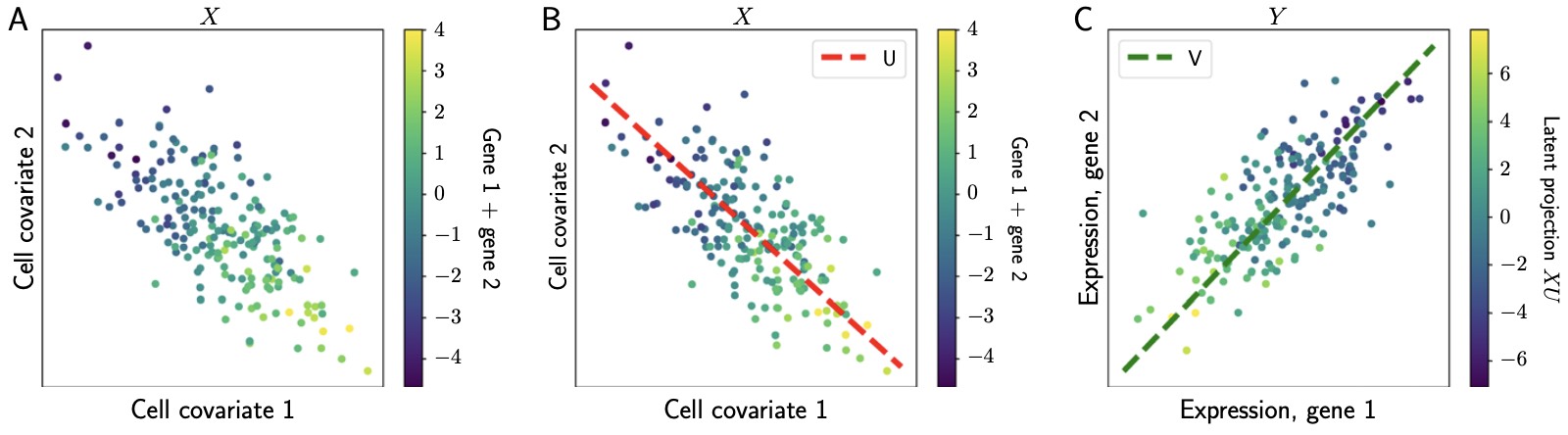
Our model, Poisson reduced rank regression (PRRR), is a Poisson regression model with a low-rank coefficient matrix.
We find that PRRR is useful in several applications. For example, finding transcriptional hallmarks of cell types (here, in scRNA-seq data from human pancreas).
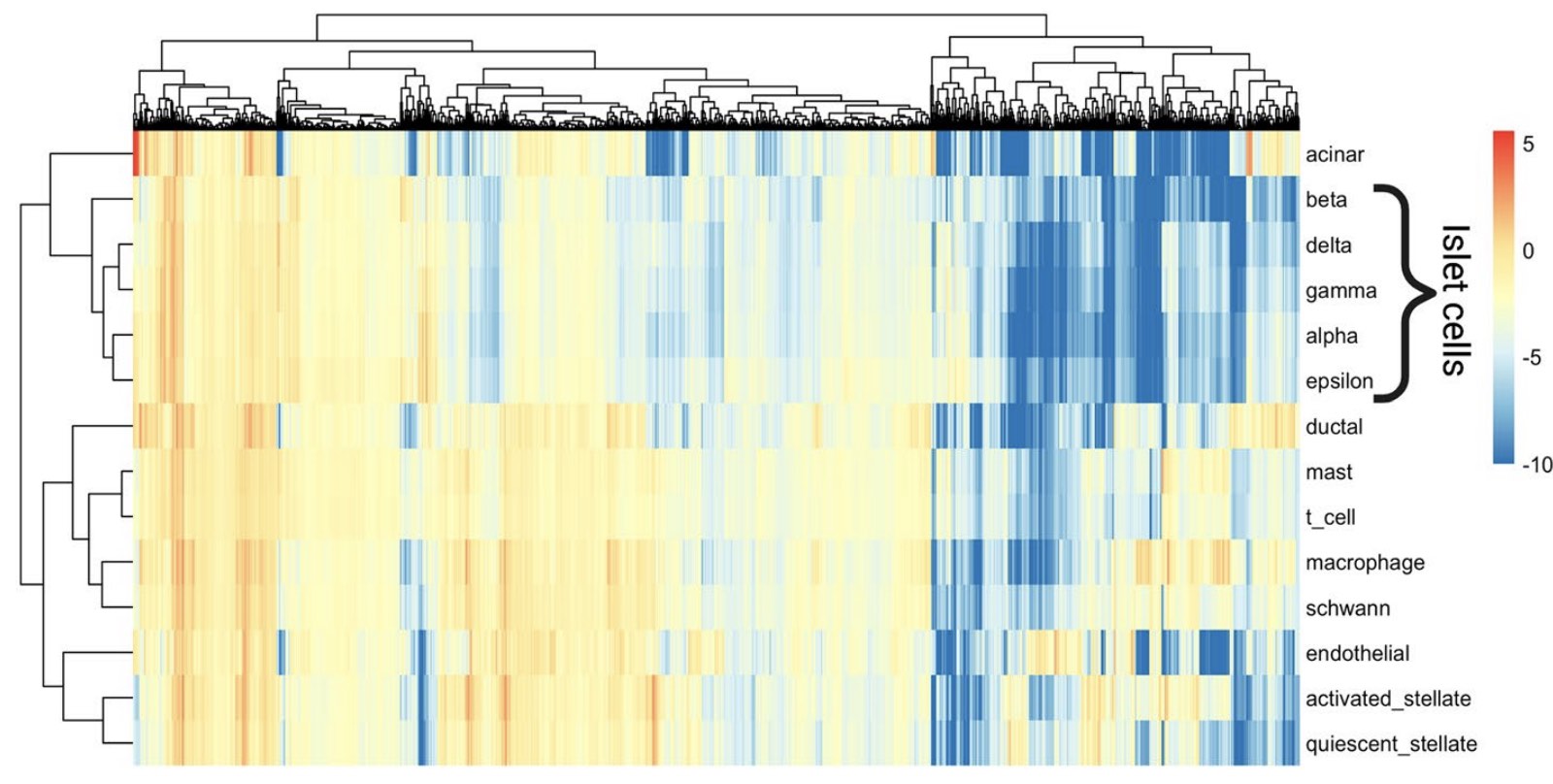
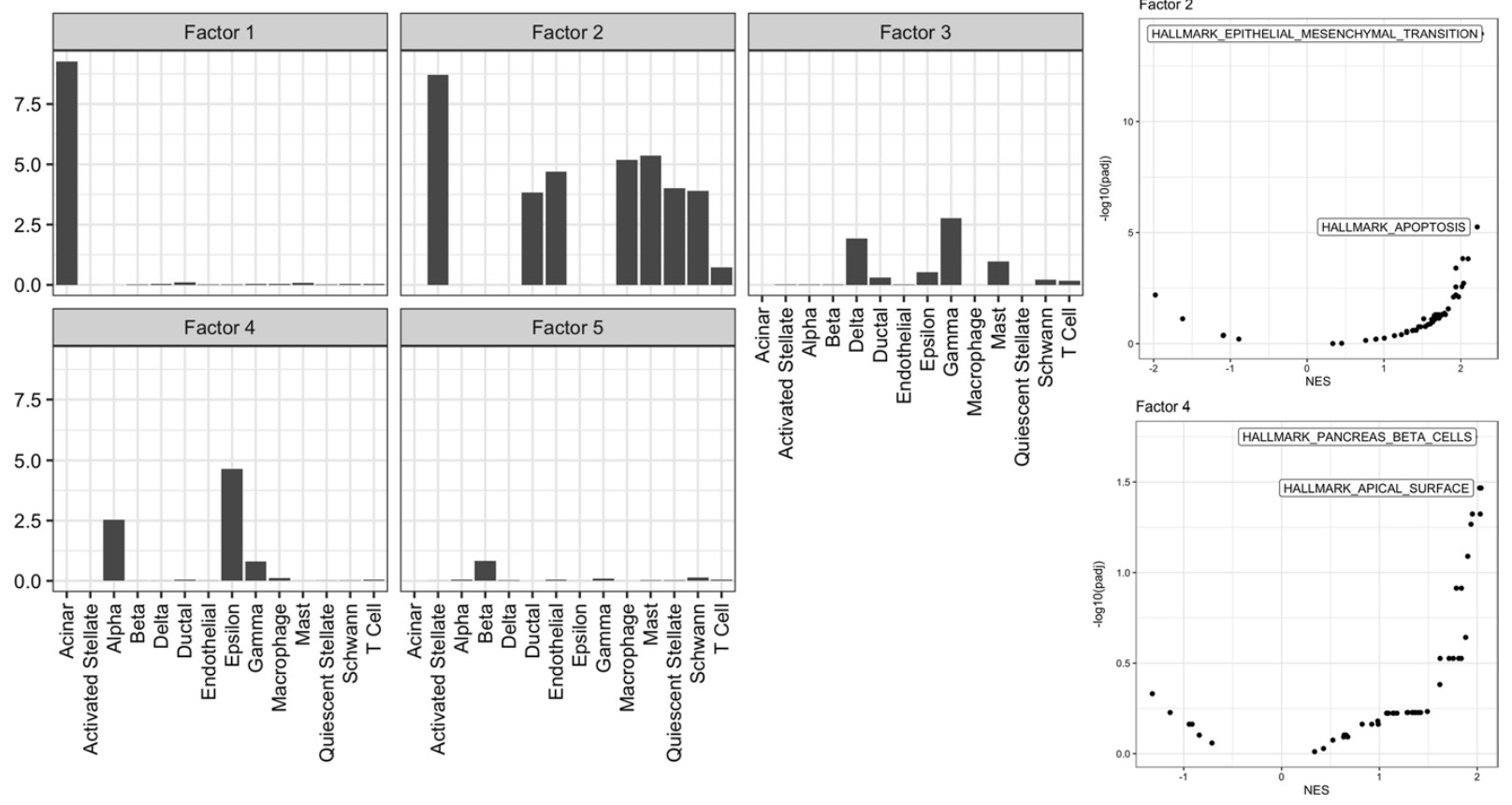
In characterizing pancreatic cell types, we find that PRRR’s learned latent factors correspond to well-established biological processes in pancreas islet and non-islet cells.
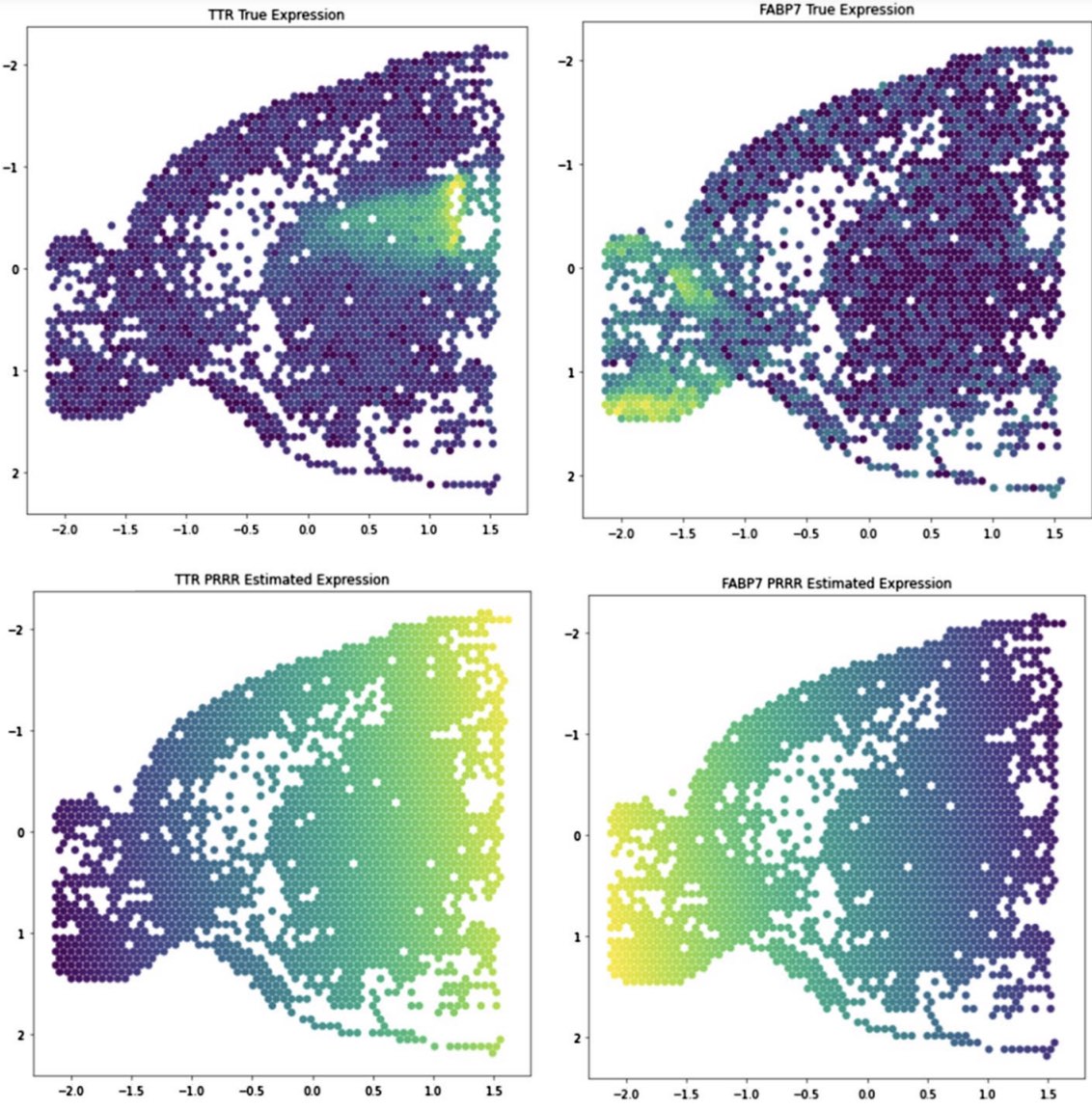
We also applied PRRR to spatial transcriptomics data, where we find that the model can identify associations between spatial coordinates and gene expression.
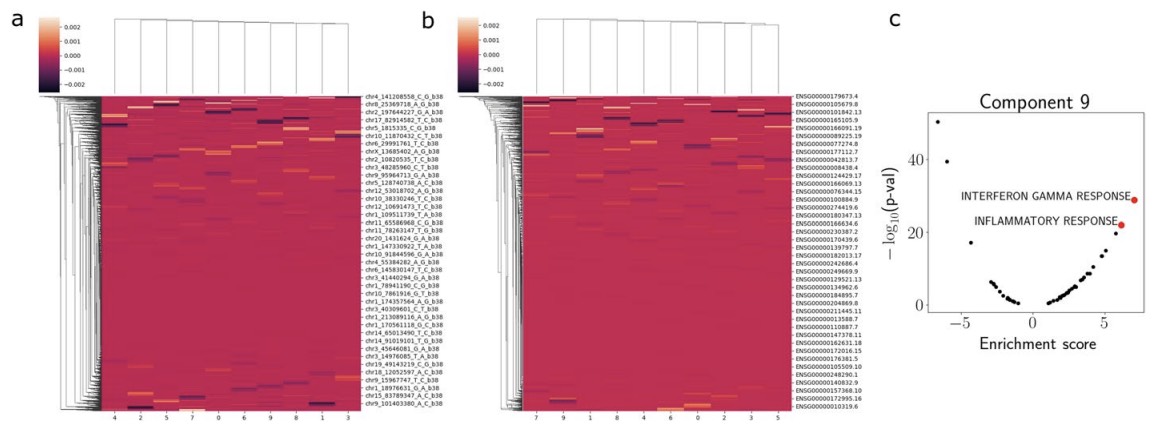
As a final application, we used PRRR to perform low-rank eQTL mapping in bulk RNA-seq data from GTEx. Applied to liver tissue samples, we find that the model’s latent factors pick up correlated eQTLs corresponding to interferon gamma and inflammatory responses.
Code for the model and experiments in the paper is here. Try it out and let us know what you think!